- Case-Based Roundtable
- General Dermatology
- Eczema
- Chronic Hand Eczema
- Alopecia
- Aesthetics
- Vitiligo
- COVID-19
- Actinic Keratosis
- Precision Medicine and Biologics
- Rare Disease
- Wound Care
- Rosacea
- Psoriasis
- Psoriatic Arthritis
- Atopic Dermatitis
- Melasma
- NP and PA
- Skin Cancer
- Hidradenitis Suppurativa
- Drug Watch
- Pigmentary Disorders
- Acne
- Pediatric Dermatology
- Practice Management
- Prurigo Nodularis
- Buy-and-Bill
News
Article
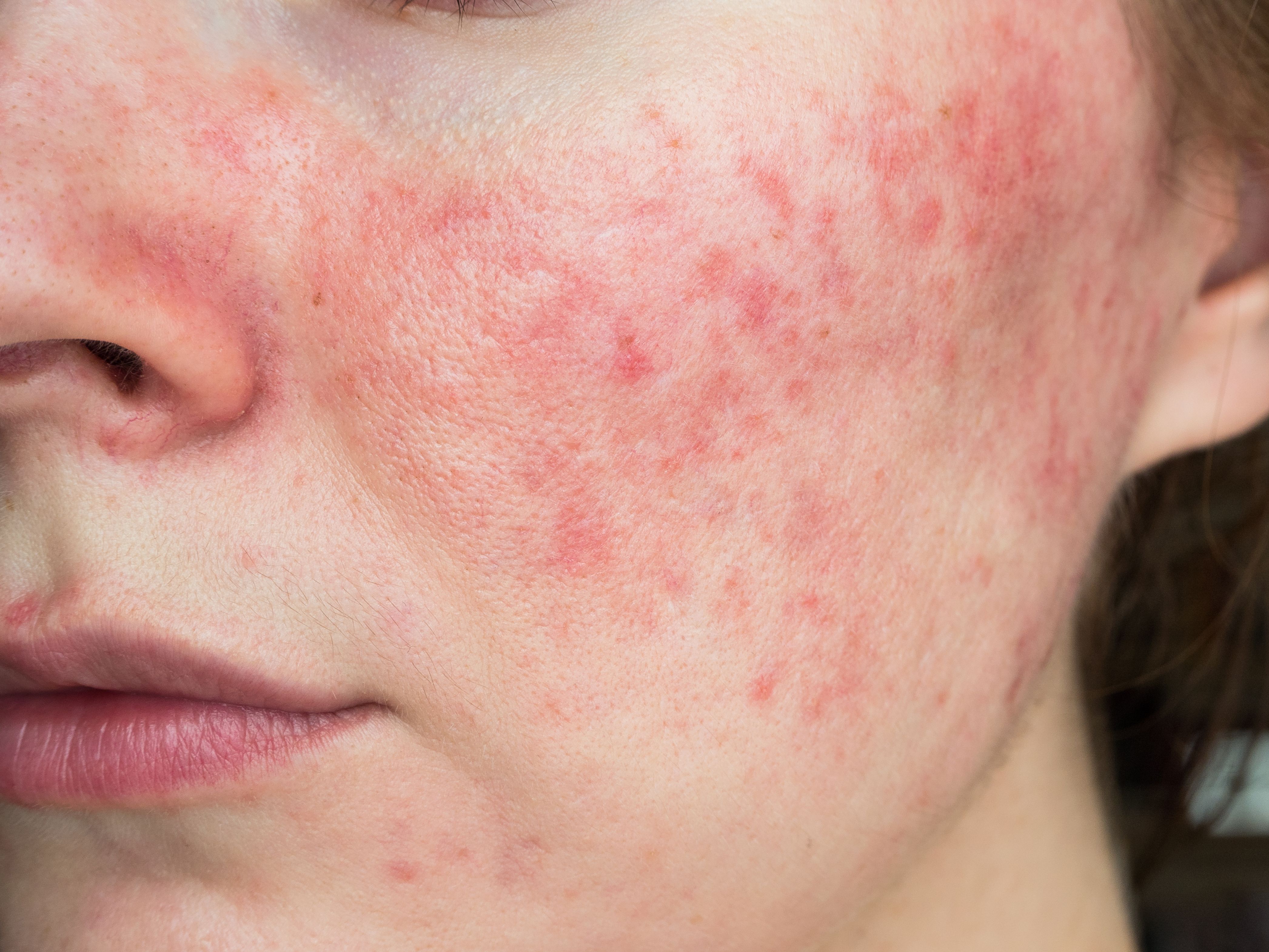
Study Links Genetic Markers in Rheumatoid Arthritis and Rosacea
Author(s):
The study found 277 differentially expressed genes shared by RA and rosacea, opening up opportunities for treatment research.
Image Credit: ©Alessandro Grandini - stock.adobe.com

Rheumatoid arthritis (RA) is characterized by persistent inflammation and synovial hyperplasia, influenced by genetic factors such as the human leukocyte antigen (HLA) system, cytokines and chemokines. The role of immune cells in RA's development and progression has been found to be significant, necessitating further research into immune mechanisms.1
Similarly, rosacea causes can include immune dysregulation, genetic factors, and microorganisms.2 Studies have suggested a link between rosacea and autoimmune diseases like RA, highlighting the importance of understanding the similarities in inflammatory cell profiles between these conditions.3-4
A recent study used bioinformatics to analyze gene expression, transcriptional networks, and immune cell infiltration in RA and rosacea, aiming to deepen the understanding of their inflammatory processes and identify new therapeutic targets.5
Methods
In this study, gene expression profiles for RA and rosacea were analyzed using data from the Gene Expression Omnibus (GEO) databases (GSE12021, GSE55457 for RA; GSE6591 for rosacea). Differentially expressed genes (DEGs) were identified using the “limma” package in R software. To explore biological functions and signaling pathways, researchers performed various analyses, including Gene Ontology (GO), KEGG pathway analysis, protein–protein interaction (PPI) network analysis, and weighted gene co-expression network analysis (WGCNA). The study stated that immune cell abundance was assessed using the CIBERSORT method, and correlations between overlapping genes and immune cell signatures were calculated using Pearson coefficients. Flow cytometry (FCM) was employed to validate the abundance of immune cells in RA and rosacea. Biomarkers and their functions were further confirmed using receiver operating characteristic (ROC) analysis, enzyme-linked immunosorbent assay (ELISA), and quantitative real-time PCR (qRT-PCR).
Results
The study identified 277 DEGs common to RA and rosacea, highlighting their roles in immune and chemokine pathways. These DEGs were linked to conditions such as infection, immunosuppression, skin lesions, and juvenile rheumatoid arthritis (JRA), suggesting that inflammatory and chemokine pathways are crucial in both diseases.
Analysis of immune cell infiltration using the CIBERSORT method revealed significant differences between RA and rosacea. In RA synovial tissues, there was an increase in plasma cells, M1 macrophages, CD8+ T cells, γδ T cells, helper T cells, and memory B cells, while M2 macrophages, resting mast cells, and monocytes were decreased. Elevated M1 macrophages in RA produce pro-inflammatory cytokines and angiogenic factors, contributing to tissue damage and increased vascularity. CXCL10, a key cytokine, was up-regulated in RA and correlated with M1 macrophages, indicating its role in RA pathology.
In rosacea, the study observed higher proportions of M1 macrophages, γδ T cells, M0 macrophages, plasma cells, and memory activated CD4+ T cells, while resting DC cells, resting mast cells, CD8+ T cells, and regulatory T cells (Tregs) were reduced. Increased M1 macrophages in rosacea tissues, activated through TLRs, lead to heightened inflammatory responses. Decreased levels of TNF-α, IL-6, IL-1β, and IFN-α in rosacea were linked to reactive oxygen species production and MAPK-NF-κB signaling in keratinocytes, promoting inflammation and angiogenesis. Reduced CCL27, which affects Treg recruitment, was noted in rosacea, suggesting its role in disease progression.
The study also identified NF-κB and RelA as key transcription factors influencing both RA and rosacea pathogenesis. CXCL10 and CCL27 emerged as potential biomarkers for RA and rosacea, respectively. These findings underscore the importance of chemokines and immune cell interactions in both diseases and highlight the need for further research to validate these biomarkers and explore other contributing immune cell subtypes.
Conclusion
Overall, the study found that rosacea and RA share common genetic markers, which could enable new molecular marker screening methods in clinical settings. Researchers found both conditions exhibit similar immune cell infiltration, particularly increased M1 macrophages. CXCL10 and CCL27 were identified as potential biomarkers for RA and rosacea, respectively, offering promising avenues to conduct research for future treatments.
References
- Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis [published correction appears in Lancet. 2016 Oct 22;388(10055):1984. doi: 10.1016/S0140-6736(16)30794-2]. Lancet. 2016;388(10055):2023-2038. doi:10.1016/S0140-6736(16)30173-8
- van Zuuren EJ. Rosacea. N Engl J Med. 2017;377(18):1754-1764. doi:10.1056/NEJMcp1506630
- Chae K, Cho M, Kim S, et al. Increased risk of rheumatoid arthritis in patients with rosacea: A nationwide population-based cohort study. J EurAcad Dermatol Venereol. 2023;37(11):e1336-e1338. doi:10.1111/jdv.19316
- Daou H, Paradiso M, Hennessy K, et al. Rosacea and the microbiome: A systematic review. Dermatol Ther (Heidelb). 2021 Feb;11(1):1-12. doi: 10.1007/s13555-020-00460-1. Epub 2020 Nov 10. PMID: 33170492; PMCID: PMC7859152.
- Wang Y, Chen J, Shen ZY, et al. Screening of diagnostic biomarkers and immune infiltration characteristics linking rheumatoid arthritis and rosacea based on bioinformatics analysis. J Inflamm Res. 2024;17:5177-5195. 2024 Aug 1. doi:10.2147/JIR.S467760