- Acne
- Actinic Keratosis
- Aesthetics
- Alopecia
- Atopic Dermatitis
- Buy-and-Bill
- COVID-19
- Case-Based Roundtable
- Chronic Hand Eczema
- Drug Watch
- Eczema
- General Dermatology
- Hidradenitis Suppurativa
- Melasma
- NP and PA
- Pediatric Dermatology
- Pigmentary Disorders
- Practice Management
- Precision Medicine and Biologics
- Prurigo Nodularis
- Psoriasis
- Psoriatic Arthritis
- Rare Disease
- Rosacea
- Skin Cancer
- Vitiligo
- Wound Care
News
Article
Dermatology Times
DermTech’s 2-GEP Assay Succeeds at Guiding Biopsy Decision-Making for Ambiguous Skin Lesions of All Skin Phototypes
Author(s):
The promising results were presented in a poster at the 2024 Winter Clinical Hawaii Dermatology Conference in Honolulu, Hawaii.
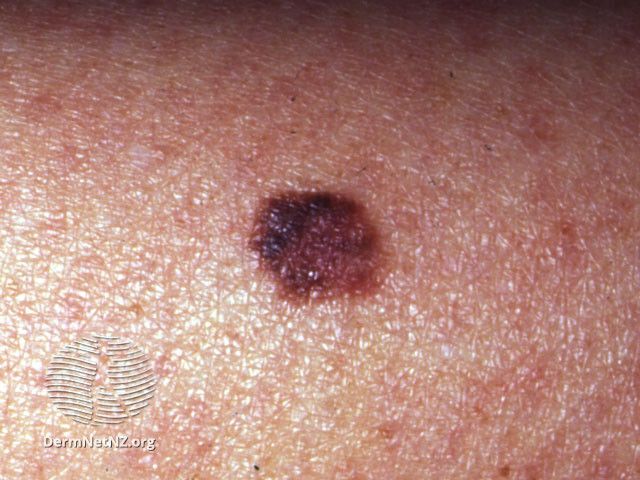
A recent poster from the 2024 Winter Clinical Hawaii Dermatology Conference in Honolulu, Hawaii, held January 12th through the 17th, analyzed the ability of DermTech’s 2-gene expression profiling (GEP) assay to detect the expression of LINC00518 and PRAME as a non-invasive assessment of clinically atypical, pigmented skin lesions to rule out melanoma with a negative predictive value (NPV) of 99%.
The non-invasive 2-GEP assay can help dermatology clinicians determine whether a biopsy is needed when a visual exam cannot rule out melanoma. For melanoma detection, “RNA extracted from skin cells collected with non-invasive adhesive patches is analyzed by RT-qPCR to detect expression of PRAME and LINC00518 RNA, 2 biomarkers that are common in melanomas but uncommon in their benign simulators.” According to Skelsey et al, LINC00518 and PRAME do not differ by skin type or ancestry, and the 2-GEP validation did not exclude any skin types. The authors noted that non-acral cutaneous melanomas in Fitzpatrick skin types IV-VI are rare and the initial 2-GEP validation study included 4 cases of patients with Fitzpatrick skin types IV-VI.
This study was conducted to better understand the real-world accuracy of the 2-GEP assay in ambiguous non-acral pigmented skin lesions in patients with Fitzpatrick skin types IV-VI and to determine whether there is a difference in results in Fitzpatrick skin types I-III.
Methods
Data collected from DermTech’s Melanoma Test Registry Protocol included patients with all Fitzpatrick skin types enrolled at 73 clinical practice locations in the US. According to the authors, most 2-GEP-negative lesions are followed by clinical surveillance and no biopsy. Therefore, the results of 2-GEP-negative lesions after follow-up examinations (unchanged/stable versus changing in a manner concerning melanoma) were recorded to track the accuracy of a negative 2-GEP result.
According to the poster, “Test performance metrics were calculated for each group (Fitzpatrick I-III and IV-VI) and groups were compared. The 95% confidence intervals for NPV and PPV were calculated using the Clopper-Pearson Exact Binomial Test, using the R function 'binom.test.' The 95% confidence intervals for the difference in NPV and PPV between the groups were calculated using the Farrington-Manning method, using the function 'farrington.manning' in R.”
2-GEP Positive and Negative Test Results
Overall, Skelsey et al’s study compared 2-GEP performance in patients with Fitzpatrick skin types (FST) I-III (n=4152) and FST IV-VI (n=130).
FST I-III
- Positive melanoma test: 66
- Positive not-melanoma test: 373
- Negative melanoma test: 4
- Negative not-melanoma test: 3709
- Sensitivity: 66/70 = 0.943
- Specificity: 3709/4082 = 0.909
FST IV-VI
- Positive melanoma test: 3
- Positive not-melanoma test: 7
- Negative melanoma test: 0
- Negative not-melanoma test: 120
- Sensitivity = 3/3 = 1.0
- Specificity = 120/127 = 0.94
NPV in FST I-III vs FST IV-VI
“The difference between the NPVs for the 2 groups was 1.0 -0.9989 = 0.0011. The 95% confidence interval for the difference in NPV between the 2 groups was -0.0299 to 0.0028 which indicates that there is no significant difference in the NPVs,” wrote Skelsey et al.
Follow-Up Days
According to the study, Fitzpatrick I-III and IV-VI group's median follow-up was 368 days and 378 days, respectively. Among the group with FST I-III and negative test results, one patient was diagnosed with melanoma in situ at their 5-month follow-up appointment. No melanomas were diagnosed in patients with FST IV-VI whose lesions tested negative.
Conclusion
In patients with FST IV-VI, all 3 melanomas diagnosed by histopathology were correctly identified by the 2-GEP assay as positive for LINC00518 and PRAME. The performance of the 2-GEP assay in patients with FST IV-VI did not differ in patients with FST I-III. According to the data, sensitivity and specificity were 90% or higher in both groups, and most importantly to the authors, the NPV for each group was greater than 99%.
“NPV is considered the most relevant metric for a rule-out test since a negative test result is often used to defer intervention (such as biopsy or excision) in favor of surveillance. During a median follow-up period of over one year, only one melanoma (in situ) was diagnosed among patients whose lesions initially tested negative, further supporting the test’s ability to appropriately guide biopsy decision-making for ambiguous pigmented skin lesions of all skin phototypes,” concluded Skelsey et al.
Reference
Skelsey M, Lofitis B, Kaufmann M, et al. Non-invasive gene expression analysis rules out melanoma with high negative predictive value regardless of skin phototype. Poster presented at: 2024 Winter Clinical Hawaii Dermatology Conference; January 12-17, 2024; Honolulu, HI.

Newsletter
Like what you’re reading? Subscribe to Dermatology Times for weekly updates on therapies, innovations, and real-world practice tips.