- Acne
- Actinic Keratosis
- Aesthetics
- Alopecia
- Atopic Dermatitis
- Buy-and-Bill
- COVID-19
- Case-Based Roundtable
- Chronic Hand Eczema
- Drug Watch
- Eczema
- General Dermatology
- Hidradenitis Suppurativa
- Melasma
- NP and PA
- Pediatric Dermatology
- Pigmentary Disorders
- Practice Management
- Precision Medicine and Biologics
- Prurigo Nodularis
- Psoriasis
- Psoriatic Arthritis
- Rare Disease
- Rosacea
- Skin Cancer
- Vitiligo
- Wound Care
Publication
Article
Dermatology Times
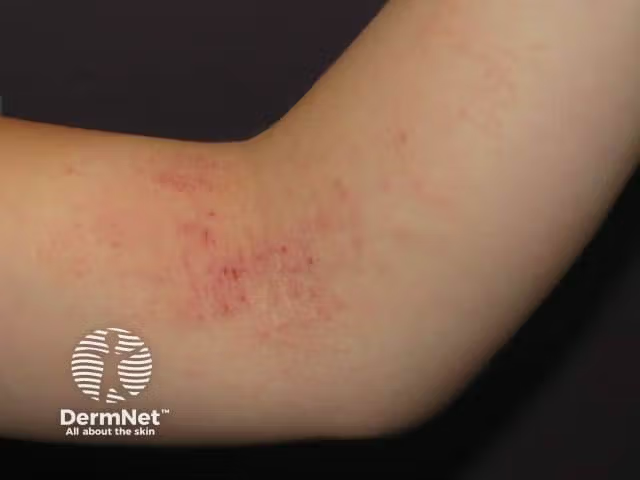
Skin microbiome as clinical biomarker for atopic dermatitis
Author(s):
With advancements in deep next-generation sequencing (NGS) and bioinformatics analysis, the authors of a recent study suggest the skin microbiome may hold promise as a clinical biomarker in atopic dermatitis (AD) management.
With advancements in deep next-generation sequencing (NGS) and bioinformatics analysis, the authors of a recent study suggest the skin microbiome may hold promise as a clinical biomarker in atopic dermatitis (AD) management.
RELATED: A basis for standardizing care for patients with atopic dermatitis
The study, published in The Journal of Allergy and Clinical Immunology, examines bacterial skin microbiome composition as well as bacterial measurement methods and technological pitfalls of using this method for AD.
“… we see promises, but also missing navigation and technological pitfalls to overcome, for the possibility of using the skin microbiome as a biomarker in AD clinical management,” they write.
According to the authors, “Skin microbiome dysbiosis, measured either as microbiome diversity or more reliably as abundance of S aureus, was clearly shown to correlate with AD severity,” noting that this may have possible prognostic value.
S Aureus levels have been shown to decrease during the treatment of AD and then return after the end of treatment in a portion of patients.
A noninvasive measurement of S aureus may become a successful treatment biomarker, indicating a sustained microbial response (SMR), and lead to a better determination of an optimal treatment.
To use this as a sustainable clinical option, however, the study authors explain that additional information is needed to select the most suitable bacterial measurement method and a standardized quantitative methodology.
RELATED: Lifestyle recommendations may curb eczema symptoms
“An important question to answer in such studies is whether the relative frequency of various bacteria (eg. S aureus frequency, as obtained from 16S-based NGS) is a good enough biomarker or whether the absolute microbial load (eg. as obtained by using quantitative PCR [qPCR]) is better,” they write.
They note that the currently relied upon microbiome sequencing method to measure the 16S gene measures the frequency of the 16S gene copies, not species frequency. New methods for measuring the real microbiome distribution need to be developed for analysis.
Various kits are used for DNA extraction from a skin sample, which leads to taxonomic selectivity bias in the yield of DNA extraction, they note.
“For example, bacteria from certain families are more easily disrupted to provide DNA through each method,” they write. “Thus, the obtained microbiome distribution is distorted through any extraction method used."
RELATED: Advances in pediatric eczema treatment
These current methods and procedures are also prone to produce differing results based on the lack of standardization in practice and can easily become contaminated from the sampling tool, extraction kit or amplification reagents, among others.
“The great disadvantage of skin sampling is the low amount of input material, in particular the low amount of bacterial DNA,” write the authors. “Furthermore, in contrast to blood or stool samples, skin sampling lacks standardization of the input material; for example, skin swab yield is highly dependent on the examiner, the material used for the swab, the applied pressure during sampling, and the size of the area sampled.”
RELATED: Topical botanicals prove effective for pediatric atopic dermatitis
Authors conclude that to further the research in this area, needs include advanced technologies and the development of a standard of practice that can be implemented in clinical trials and large longitudinal registries.
“It is critical to develop standardized quantitative methodologies for skin bacterial microbiome analysis, which can be tested, compared and validated in these studies,” authors write. “In addition, new promising technologies, such as single-molecule real-time sequencing, which could improve skin microbiome analysis with greater accuracy and/or longer sequencing length, and more advanced bioinformatic tools needs to be further developed and tested.”
References:
Reiger M, Traidl-hoffmann C, Neumann AU. The skin microbiome as a clinical biomarker in atopic eczema: Promises, navigation, and pitfalls. J Allergy Clin Immunol. 2020;145(1):93-96.